Abstract
Introduction:
We analyzed genomic profile in line with immune checkpoint molecule expression in multiple myeloma using whole genome sequencing (WGS) and whole transcriptome sequencing (WTS) data.
Methods:
We performed WGS of CD138+ selected tumor and the matched normal from saliva of multiple myeloma patients at 90-60x and 30x respectively from 37 patients. At the same time, we performed WTS using both tumor (CD138+) and immune milieu (CD138-) samples from these patients. Structural variations were called with Delly2 and Manta and the intersect of two call sets were used as the final SV calls. Copy number analysis was done with Sequenza and custom scripts. Somatic single nucleotide variants were identified with Mutect1 and Strelka and filtered against the panel of normal. RNA-seq was aligned using STAR algorithm and gene expression was quantified using RSEM algorithm. We extracted the expression of 21 immune checkpoint-related molecules (LAG3, PD-L1, IDO1, TIM3, CD27, CTLA-4, ICOS, TIGIT, PD-1, PD-L2, VISTA, and their ligands). To compare cell populations within a set of CD138-sorted MM samples, we used CIBERSORT algorithm to estimate predict fractions of leukocyte RNA.
Results & Discussion:
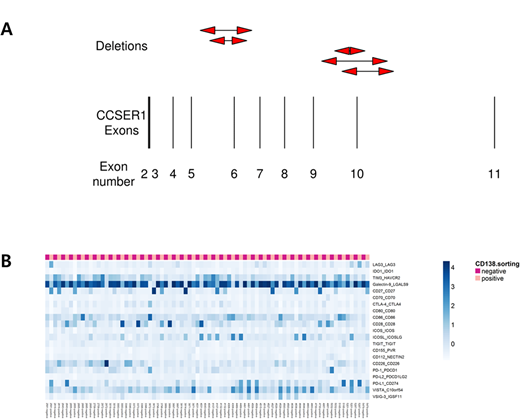
A total of 1,359 somatic structural variants were identified, with a median of 28 structural variant events per patient. The most common translocation event was t(4;14) (n=6, 16.2%). We identified novel recurrent deletions in CCSER1 (n=5, 13.5%) either deleting exon 10 (n=3) or exon 6 (n=2). CCSER1 has been proposed to a have a role in cell division with defects leading to cytokinetic defects. We observed an enrichment of whole-genome duplicated samples with CCSER1 deleted patients. We propose that CCSER1 deletion may have a role in whole-genome duplication events seen in multiple myeloma. Chromothripsis was also seen in 7 patients (18.9%). A total of 9 chromothripsis events were observed and chromosome 6 was most frequently involved (n=3). Recurrent mutation in IGKV4-1 chr2: 89185484 and 89185485 (p.Ala60) was observed suggesting dysfunctional antigen recognition in myeloma cells.
From immune checkpoint perspective, expression of Galectin-9, a ligand of TIM3, was outstandingly high in immune milieu suggesting potential role of TIM3 inhibitor in myeloma. On the other hand, VISTA was highly expressed in tumors. When we matched the transcriptomic profiles with genomic variants, NRAS G61R mutant showed high expression of TIM3/Galectin-9 axis suggesting characteristics immune milieu of RAS mutant myeloma.
Conclusions:
We could identify novel CCSER1 recurrent deletion and IGKV4-1 mutation in multiple myeloma using WGS. WTS immune milieu analysis suggest potential role of TIM3 inhibitor in myeloma, especially in RAS mutated cases.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal